Released datasets and visualizations in Neuroglancer
The current released segmentation is FAFB-FFN1-20200412, freely viewable for exploration and circuit tracing.
This reconstruction is based on the Full Adult Fly Brain (FAFB) dataset. Note that the IDs in this release should be consistent with IDs in the previous 20190805 release.
Sampling of reconstructed neurons
(For additional examples with Neuroglancer links, see the Gallery.)
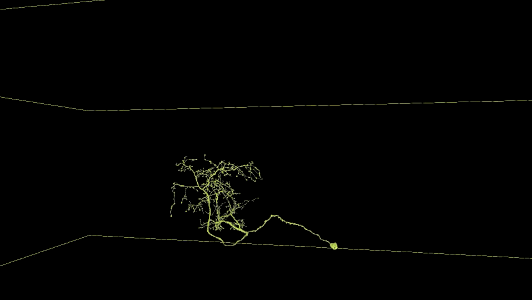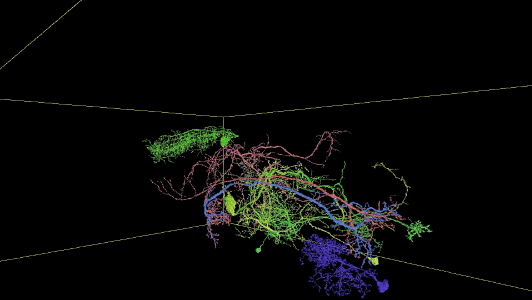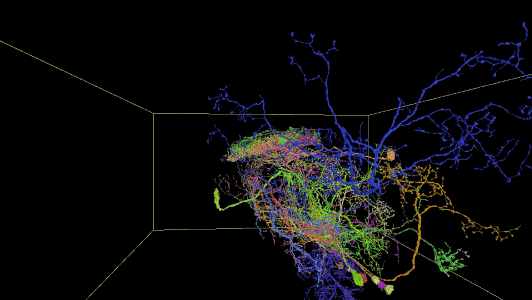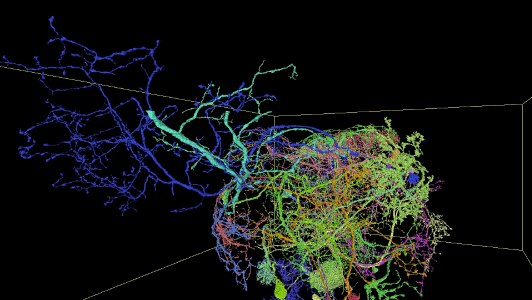
Description of Neuroglancer layers
- fafb_v14: the raw imagery of the publicly available Full Adult Fly Brain (FAFB) described by Zheng et al., 2018, in the v14 global alignment.
- fafb_v14_clahe: as above, with CLAHE post-processing.
- fafb-ffn1-20200412: the current public automated segmentation, produced by Flood-Filling Networks (FFNs) with added procedures for handling common serial-section EM artifacts. 3d meshes and TEASAR-based automated skeletonization are also included. See manuscript for details; updates pending.
- synapses_Buhmann2019: synapse pre/post partner annotations from Buhmann et al., 2019.
- clefts_Heinrich_etal: synaptic cleft mask predictions from Heinrich et al., 2018.
- neuropil-regions-surface: 3d surface meshes for canonical fly brain regions of interest; details and label mapping described here. To see the region labels, right click the layer name.
- neuropil-full-surface: 3d surface mesh for the entire canonical fly brain also described at link above.
- public_skeletons: the publicly released manually traced skeletons from Zheng et al., 2018, in the v14 global alignment. To see the neuron type labels, right click the layer name.
The web viewer is Neuroglancer; see GitHub for documentation, or use the upper-right question mark button for quick tips.
Demo using Neuroglancer to explore Olfactory Projection Neurons and Kenyon Cells of the Mushroom Body in the earlier 20190521 release:
Additional data
Besides the data in the combined view layers above, we also provide:
- Tissue mask volume used to restrict FFN segmentation movement and to compute the full brain mesh and number of segmentable voxels. This version is represented as a semantic segmentation with each voxel assigned to its highest probability class. In FFN segmentation, we used raw class probabilities to control movement; see manuscript. Class labels are:
- neuropil
- soma
- glia
- empty
- resin
- tissue border
- Section-to-section cross-correlation volume used for Local Realignment, Irregular Section Substitution, and fallback Movement Restriction. This gives the detected shifts between section Z and Z+1, as computed by patch-wise section-to-section cross-correlation and peak-finding. Shift values are given in # of 32 nm voxels. In the Neuroglancer view, the RGB channel values encode X-shift (red), Y-shift (green), and peak correlation value (blue) which is used as patch match score.
Data access from Python via CloudVolume
Interactive access to the segmentation from Python is easy via CloudVolume. Basic access is demonstrated in this Colab notebook. Since looking up segment IDs at a series of points throughout the volume is a common use case, this notebook also demonstrates a method for batching point lookups to more efficiently access storage chunks.
Download links and format
Data can also be downloaded directly from Google Cloud Storage (e.g. via gsutil) from the links listed in Neuroglancer, e.g. for the segmentation: gs://fafb-ffn1-20200412/segmentation. The format specification for the data is described here.
Note that the Heinrich clefts n5 data set has an offset of 1 z-section; this is corrected in the Neuroglancer views, but not in the underlying dataset.
64-bit TIFF image tiles of the previous 2019-08-05 segmentation at 16x16x40 nm resolution (1 image per section) can be downloaded at gs://fafb-ffn1-20190805/segmentation_tiffs/agglomerated_flat_16nm.