Automated Reconstruction of a Serial-Section EM Drosophila Brain with Flood-Filling Networks and Local Realignment
The Connectomics at Google team and our academic collaborators have publicly released a segmentation of an entire fly brain for use in neuroscience and image processing research. We have also released an updated methods manuscript to accompany the dataset.
Released datasets and visualizations in Neuroglancer
The current released segmentation is FAFB-FFN1-20190805, freely viewable for exploration and circuit tracing.
Sampling of neurons reconstructed in the released segmentation:
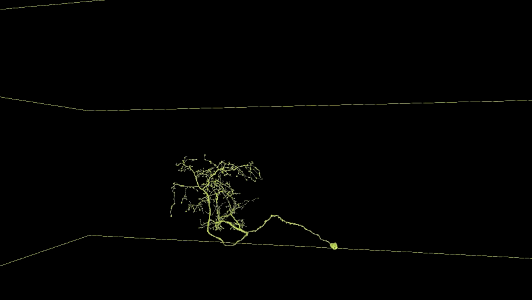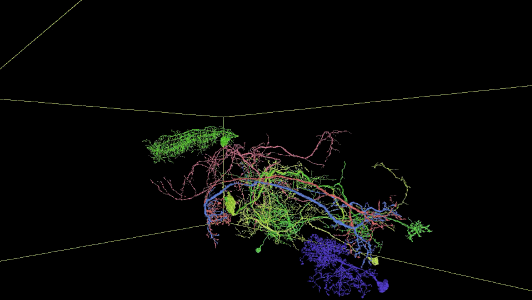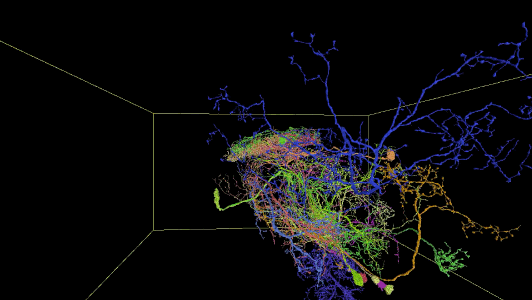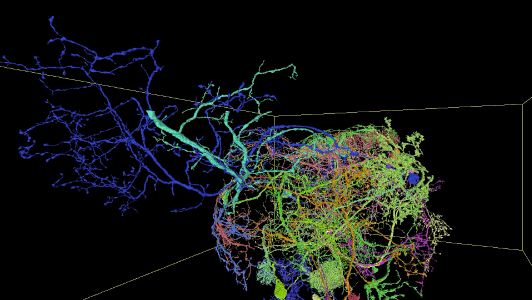
Description of Neuroglancer layers
- fafb_v14: the raw imagery of the publicly available Full Adult Fly Brain (FAFB) described by Zheng et al., 2018, in the latest v14 global alignment.
- fafb_v14_clahe: as above, with CLAHE post-processing.
- fafb-ffn1-20190805: the current public automated segmentation, produced by Flood-Filling Networks (FFNs) with added procedures for handling common serial-section EM artifacts. See manuscript for details.
- synapses_Buhmann2019: synapse pre/post partner annotations from Buhmann et al., 2019.
- clefts_Heinrich_etal: synaptic cleft mask predictions from Heinrich et al., 2018.
- neuropil-regions-surface: 3d surface meshes for canonical fly brain regions of interest; details and label mapping described here.
- neuropil-full-surface: 3d surface mesh for the entire canonical fly brain also described at link above.
- skeletons_32nm: automated TEASAR skeletonization of the FAFB-FFN1-20190805 segmentation.
- public_skeletons: the publicly released manually traced skeletons from Zheng et al., 2018, in the latest v14 global alignment. Skeleton IDs from the valid ID list must be entered manually to select which IDs to display. See here for a simplified view with all public skeletons active.
The web viewer is Neuroglancer; see GitHub for documentation, or use the upper-right question mark button for quick tips.
Demo using Neuroglancer to explore Olfactory Projection Neurons and Kenyon Cells of the Mushroom Body in the previous 20190521 release:
Additional data
Besides the data in the combined view layers above, we also provide:
- Tissue mask volume used to restrict FFN segmentation movement and to compute the full brain mesh and number of segmentable voxels. This version is represented as a semantic segmentation with each voxel assigned to its highest probability class. In FFN segmentation, we used raw class probabilities to control movement; see manuscript. Class labels are:
- neuropil
- soma
- glia
- empty
- resin
- tissue border
- Section-to-section cross-correlation volume used for Local Realignment, Irregular Section Substitution, and fallback Movement Restriction. This gives the detected shifts between section Z and Z+1, as computed by patch-wise section-to-section cross-correlation and peak-finding. Shift values are given in # of 32 nm voxels. In the Neuroglancer view, the RGB channel values encode X-shift (red), Y-shift (green), and peak correlation value (blue) which is used as patch match score.
Data access from Python via CloudVolume
Interactive access to the segmentation from Python is easy via CloudVolume. Basic access is demonstrated in this Colab notebook. Since looking up segment IDs at a series of points throughout the volume is a common use case, this notebook also demonstrates a method for batching point lookups to more efficiently access storage chunks.
Download links and format
Data can also be downloaded directly from Google Cloud Storage (e.g. via gsutil) from the links listed in Neuroglancer, e.g. for the segmentation: gs://fafb-ffn1-20190805/segmentation. The format specification for the data is described here.
64-bit TIFF image tiles of the segmentation at 16x16x40 nm resolution (1 image per section) can be downloaded at gs://fafb-ffn1-20190805/segmentation_tiffs/agglomerated_flat_16nm.
Manuscript
- The accompanying manuscript describing our methodology, validation experiments, and released resources is posted on bioRxiv.
- Abstract:
Reconstruction of neural circuitry at single-synapse resolution is an attractive target for improving understanding of the nervous system in health and disease. Serial section transmission electron microscopy (ssTEM) is among the most prolific imaging methods employed in pursuit of such reconstructions. We demonstrate how Flood-Filling Networks (FFNs) can be used to computationally segment a forty-teravoxel whole-brain Drosophila ssTEM volume. To compensate for data irregularities and imperfect global alignment, FFNs were combined with procedures that locally re-align serial sections as well as dynamically adjust and synthesize image content. The proposed approach produced a largely merger-free segmentation of the entire ssTEM Drosophila brain, which we make freely available. As compared to manual tracing using an efficient skeletonization strategy, the segmentation enabled circuit reconstruction and analysis workflows that were an order of magnitude faster.
Changelog
- 2020-01-21: Buhmann et al. synapse predictions added.
- 2019-11-06: section-to-section cross-correlation volume released.
- 2019-11-01: tissue mask volume released.
- 2019-09-30: notebook demonstrating CloudVolume access released, public skeletons added to Neuroglancer instance.
- 2019-08-20: automated skeletonization added to Neuroglancer instance, TIFF images of segmentation available for download.
- 2019-08-05: updated segmentation and manuscript release.
- 2019-06-19: initial posting.

